Frequently Asked Questions
Is there any contaminating RNA in the DNA obtained using the DNAstorm™ kit?
Contamination from RNA is eliminated by performing an optimized RNase digestion step immediately following the lysis step.
How much DNA can I expect to obtain from an FFPE sample?
The biggest variable that affects the total amount of DNA obtained is the quality of the sample itself (i.e. the type and amount of tissue, and the care taken in isolation and preservation of the sample). Using the DNAstorm™ kit, and assuming at least reasonable sample quality, amounts greater than 1 µg can be obtained.
Can DNA obtained using the DNAstorm™ kit be used in next-generation sequencing?
Yes. Good quality libraries can be obtained, providing that the DNA is of sufficiently high quality.
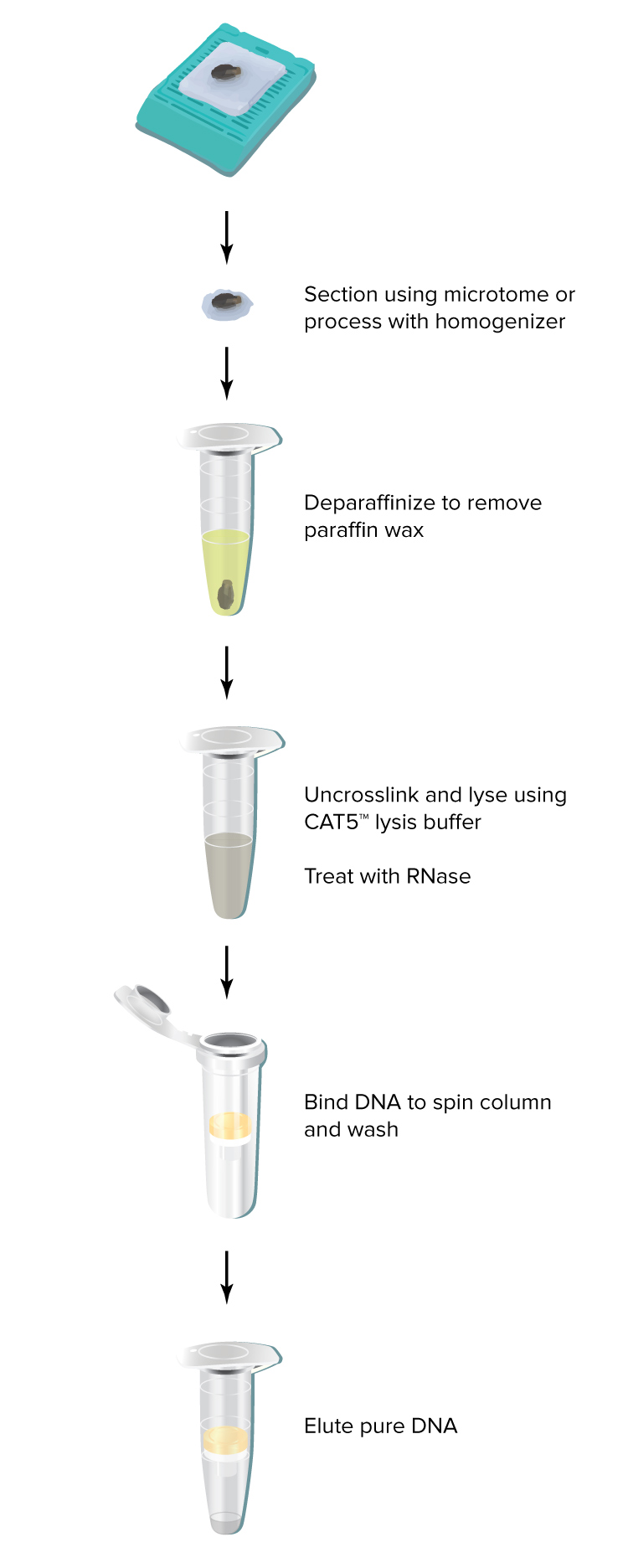
How should the tissue be prepared?
Use a microtome to obtain 5-10 µm sections from FFPE samples. Sections thinner than 5 µm may be used if they can be reliably cut. Sections thicker than 10 µm are not recommended because they may not be fully digested.
Can I use tissue that is not paraffin-embedded?
Yes, tissue can be used which is not embedded in paraffin. In this case, we recommend mechanically grinding an amount of tissue equivalent to the recommended number of sections.
Can I use FFPE cores?
Yes, FFPE cores can be used. Because cores are not processed using a microtome, sample digestion tends to be more difficult and mechanical homogenization (e.g. using steel beads) is recommended if incomplete digestion is observed.
Which deparaffinization method do you recommend?
The DNAstorm™ kit includes a recommended Deparaffinization Reagent. Unlike other common methods (e.g. xylenes), the Deparaffinization Reagent is efficient, non-toxic and does not require the use of a fume hood. In our testing, the included reagent is at least as effective as xylenes at removing paraffin and allowing purification of high quality nucleic acids.
After mixing the Deparaffinization Reagent with the CAT5 Lysis Buffer, I see a white cloudy layer in between the Deparaffinization Reagent layer and the aqueous layer. What is this and how does it affect the extraction?
The white cloudy layer is an emulsion between the Deparaffinization Reagent and the CAT5 Lysis Buffer, which may form when these two reagents are vortexed or given a hard mix. To avoid this issue, we recommend not vortexing the sample when the Deparaffinization Reagent and CAT5 Lysis Buffer are in contact. When mixing is necessary in the presence of both these reagents (e.g. when protease is added), we recommend pipette mixing. The white cloudy layer can be removed by giving the sample a hard spin at maximum speed (> 16,000 x g) for at least 2 minutes. The length of time will depend on the volume of the emulsion.
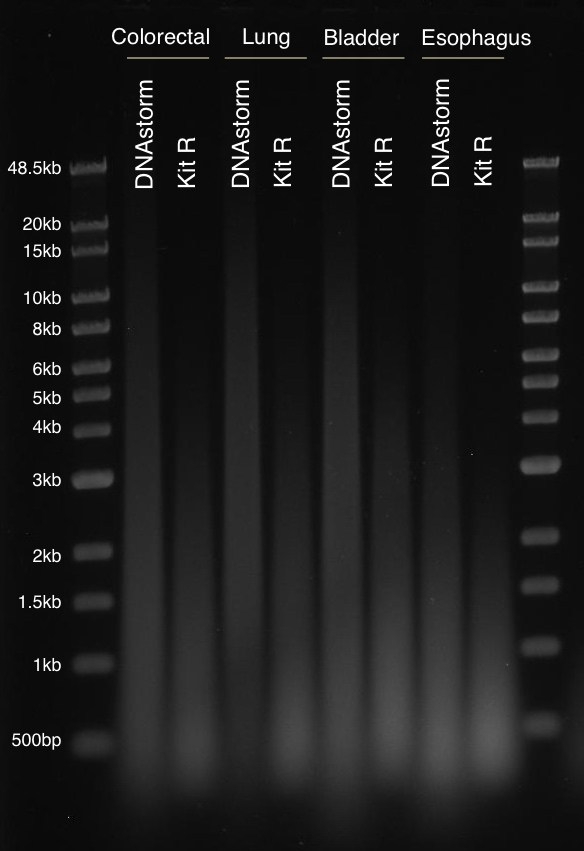
How can I evaluate the integrity of the DNA I obtained?
Due to the wide size distribution of DNA isolated from FFPE tissue samples, we recommend using pulsed-field gel electrophoresis (PFGE). Methods based on capillary electrophoresis such as the Agilent BioAnalyzer can also be used, but may not properly resolve high molecular weight fragments (greater than 10k) in better-quality samples.
Why does my extracted DNA fail to amplify properly? I notice a lot of PCR inhibition and/or Ct values that make no sense.
PCR inhibition is often observed when high amounts of FFPE-extracted template DNA are used. The inhibition is usually not due to the presence of contaminants, but results from residual chemical modifications and damage in the DNA itself. Several simple adjustments to the PCR protocol can overcome this issue. First, the amount of template DNA should be decreased. Second, the amount of PCR polymerase should be increased by 2-4x. Third, the annealing and extension times should be extended. Fourth, the amount of dNTPs can be increased.
An in-depth discussion of this issue is found in Dietrich et al. (2013), PLoS ONE 8(10): e77771.